Projects
GUDMAP/RBK Analytics

Design and manage the Agile group responsible for creating analytics for data stored on the NIH consortiums GUDMAP and RBK. This analytics are not only best-practices pipelines written in Nextflow, but also integrates seamlessly with the consortiums' data-hub, in terms of initiation, and data ingress/egress. It is built with the flexibility to utilize on premises HPC, or a custom built AWS architecture (including many serverless resources) for low-cost, highly-available queueing, compute, and reporting. It can also be easily adapted to almost any running environment using custom configs. The entire pipeline utilizes dockerized micro-services which is orchestrated by Nextflow.
Nextflow in the Cloud
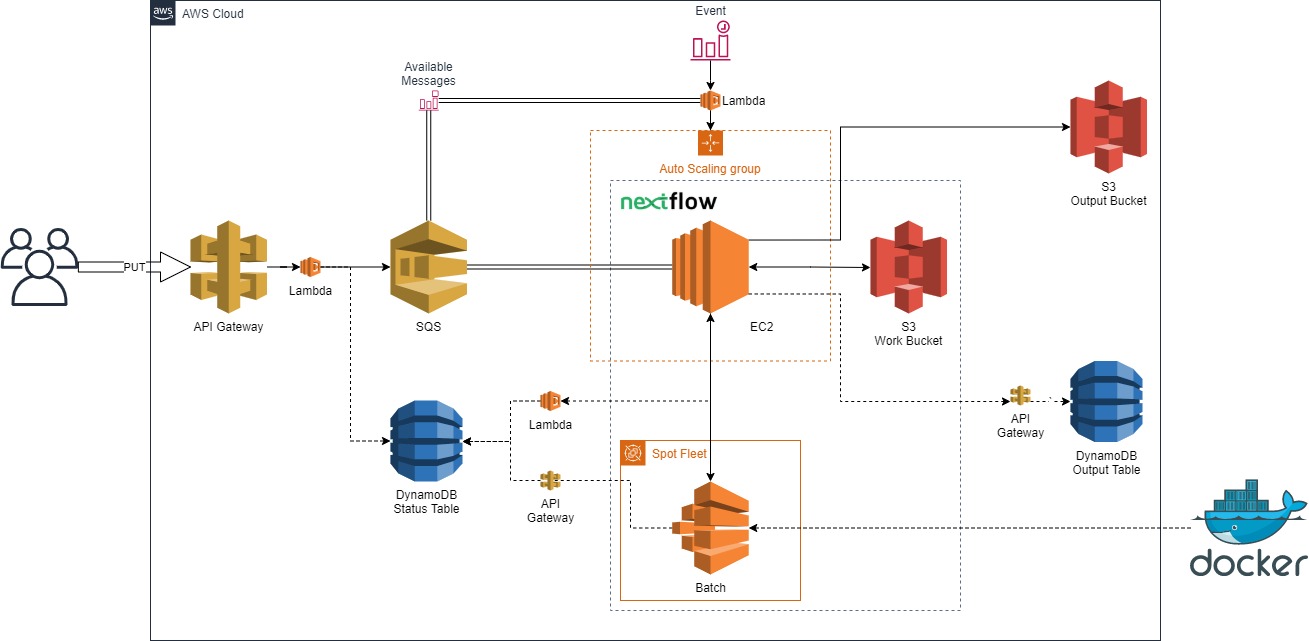
Manage a small agile team to create a proof of concept AWS architecture to run Nextflow pipelines in the cloud. It utilizes low-cost, highly-availability queueing, compute, and storage resources. (NOTE: project decomissioned and the git repository is no longer public, for more information feel free to reach out to [Gervaise Henry](ghenry@gmail.com))
Pipeline Run Tracking
Creating a pipeline run tracking tool for the Bioinformatics Core Facility at UT Southwestern Medical Center. This tool utilizes AWS serverless offerings, including REST-API and a website for run tracking. (NOTE: project decomissioned and the git repository is no longer public, for more information feel free to reach out to [Gervaise Henry](ghenry@gmail.com))
Strand Lab External Website
The Strand Lab website was designed to not only provide a location to share information about the lab, but also provide a means to showcase the lab's data and biorepository. The single-cell data is displayed using CZI's cellxgene visualization tool as well as pre-generated genome-wide visualization of the expression data. The lab has an extensive biorepository of human lower urinary tract biosamples and de-identified clinical data as well as H&E images are available for viewing and filtering on the website. The site is hosted serverlessly on AWS.
Strand Lab Internal Data Website

Developed the internal Strand Lab website and manage the server used explore and share pre-publication single-cell RNA-sequencing data. (NOTE: project is private, for more information feel free to reach out to [Gervaise Henry](ghenry@gmail.com))
BICF Pipelining

Creating analytics for the Bioinformatics Core Facility at UT Southwestern Medical Center, using the pipelining language Nextflow.